The function takes a list of promotions, original width, and original height as input, and returns an array of decoded output masks.
153 {
154 ZCT c = App.viewer.GetCoordinate();
155 List<float[]> lts = (List<float[]>)b.Tag;
156 int fr = b.GetFrameIndex(c.Z, c.C, c.T);
157 var embedding_tensor = new DenseTensor<float>(lts[fr], new[] { 1, 256, 64, 64 });
158 var bpmos = promotions.FindAll(e => e.mType == PromotionType.Box);
159 var pproms = promotions.FindAll(e => e.mType == PromotionType.Point);
160 int boxCount = promotions.FindAll(e => e.mType == PromotionType.Box).Count();
161 int pointCount = promotions.FindAll(e => e.mType == PromotionType.Point).Count();
162 float[] promotion = new float[2 * (boxCount * 2 + pointCount)];
163 float[] label = new float[boxCount * 2 + pointCount];
164 for (int i = 0; i < boxCount; i++)
165 {
166 var input = bpmos[i].GetInput();
167 for (int j = 0; j < input.Count(); j++)
168 {
169 promotion[4 * i + j] = input[j];
170 }
171 var la = bpmos[i].GetLable();
172 for (int j = 0; j < la.Count(); j++)
173 {
174 label[2 * i + j] = la[j];
175 }
176 }
177 for (int i = 0; i < pointCount; i++)
178 {
179 var p = pproms[i].GetInput();
180 for (int j = 0; j < p.Count(); j++)
181 {
182 promotion[boxCount * 4 + 2 * i + j] = p[j];
183 }
184 var la = pproms[i].GetLable();
185 for (int j = 0; j < la.Count(); j++)
186 {
187 label[boxCount * 2 + i + j] = la[j];
188 }
189 }
190
191 var point_coords_tensor = new DenseTensor<float>(promotion, new[] { 1, boxCount * 2 + pointCount, 2 });
192
193 var point_label_tensor = new DenseTensor<float>(label, new[] { 1, boxCount * 2 + pointCount });
194
195 float[] mask = new float[256 * 256];
196 for (int i = 0; i < mask.Count(); i++)
197 {
198 mask[i] = 0;
199 }
200 var mask_tensor = new DenseTensor<float>(mask, new[] { 1, 1, 256, 256 });
201
202 float[] hasMaskValues = new float[1] { 0 };
203 var hasMaskValues_tensor = new DenseTensor<float>(hasMaskValues, new[] { 1 });
204
205 var decode_inputs = new List<NamedOnnxValue>();
206
207 float[] orig_im_size_values = { (float)orgHei, (float)orgWid };
208 var orig_im_size_values_tensor = new DenseTensor<float>(orig_im_size_values, new[] { 2 });
209 decode_inputs = new List<NamedOnnxValue>
210 {
211 NamedOnnxValue.CreateFromTensor("image_embeddings", embedding_tensor),
212 NamedOnnxValue.CreateFromTensor("point_coords", point_coords_tensor),
213 NamedOnnxValue.CreateFromTensor("point_labels", point_label_tensor),
214 NamedOnnxValue.CreateFromTensor("mask_input", mask_tensor),
215 NamedOnnxValue.CreateFromTensor("has_mask_input", hasMaskValues_tensor),
216 NamedOnnxValue.CreateFromTensor("orig_im_size", orig_im_size_values_tensor)
217 };
218
219 var segmask = this.decoder.Run(decode_inputs);
220 var outputmask = segmask.First().AsTensor<float>().ToArray();
221 BioLib.Recorder.AddLine("App.samTool.microsam.Decode(Images.GetImage(\"" + b.Filename + "\"),App.samTool.Promotions," + orgWid + "," + orgHei + ");", false);
222 return outputmask;
223 }

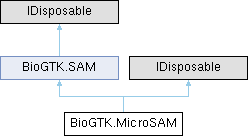
 Public Member Functions inherited from BioGTK.SAM
Public Member Functions inherited from BioGTK.SAM Static Public Member Functions inherited from BioGTK.SAM
Static Public Member Functions inherited from BioGTK.SAM Public Attributes inherited from BioGTK.SAM
Public Attributes inherited from BioGTK.SAM Static Public Attributes inherited from BioGTK.SAM
Static Public Attributes inherited from BioGTK.SAM